
My PhD research focuses on understanding evolutionary dynamics of adaptation of complex traits using Drosophila melanogaster as a model organism. I work with a study system that was created in the 80s, composed of a set of populations that are on a selection treatment for postponed reproduction - we only allow old flies to reproduce. That generates some interesting phenotypes, such as flies with more than double the regular lifespan. Right now, I am focused at the genomics of that adaptation. Hopefully, in understanding the evolutionary dynamics that happened in this system, we can understand a bit more about some fundamental mechanisms of evolution, such as its reproducibility, reversibility, and parallelity.
Previous research#
iPSC Pipeline#
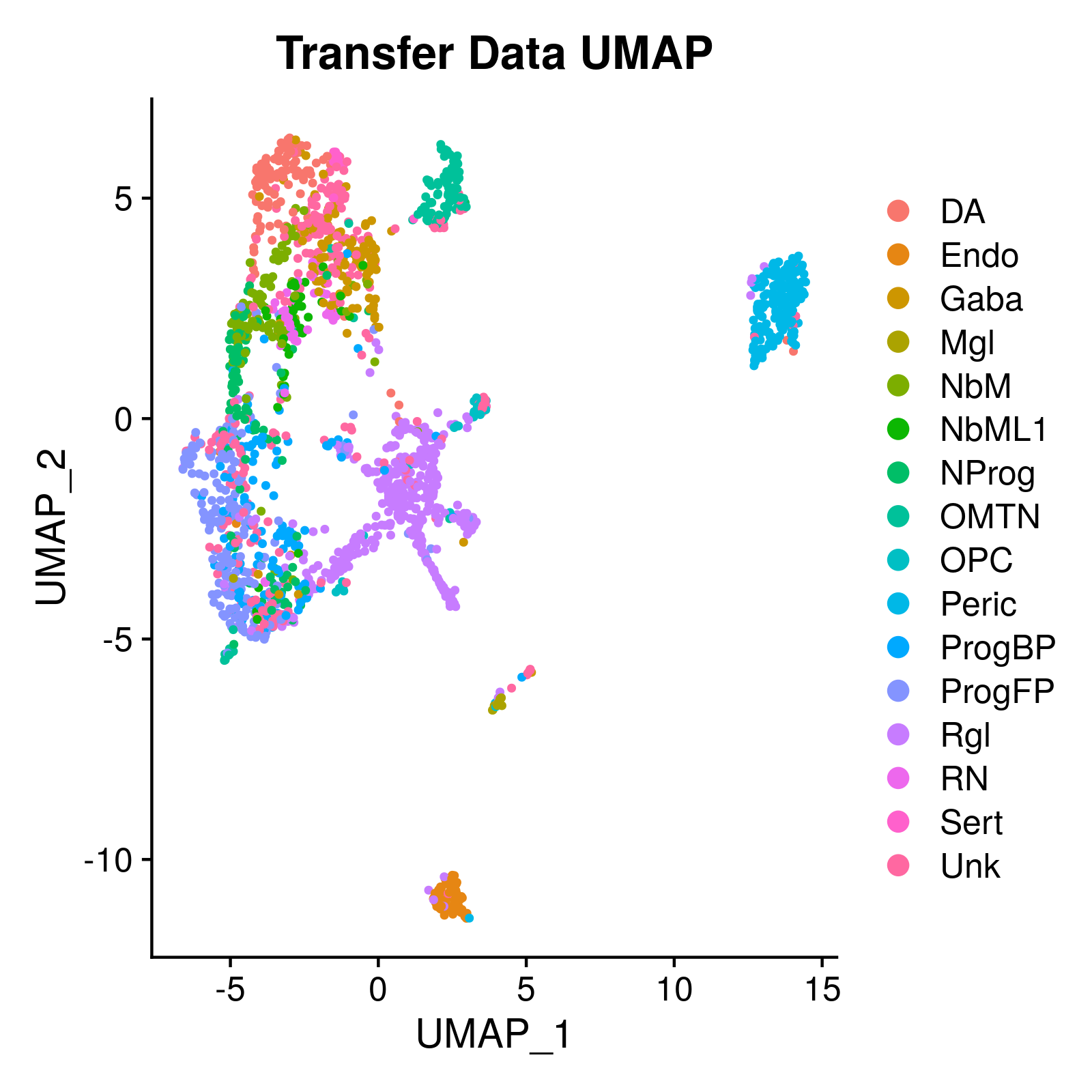
As an undergrad researcher at the Federal University of Santa Catarina, I participated in several projects and playing different roles, from the bioinformatician to the field biologist. My most successful project was the development of a bioinformatics workflow to analyise single-cell iPSC RNA-Seq data using a selection of tools to generate different reports. That workflow uses Snakemake and runs on a Docker image. This project was critical for my understanding of both tools, and getting my bioinformatics game to the next level.
Dichotomous Key App#
I also developed a HTML/Javascript app to navigate dichotomous keys digitally. Both undergraduate and graduate students use it in systematics classes. That project improved my undertanding of coding and understanding the challenge of putting a simple intellectual algorithm into an app that people will use.
